Cell Locator: Manually align specimens to annotated 3D spaces
Intro and Summary
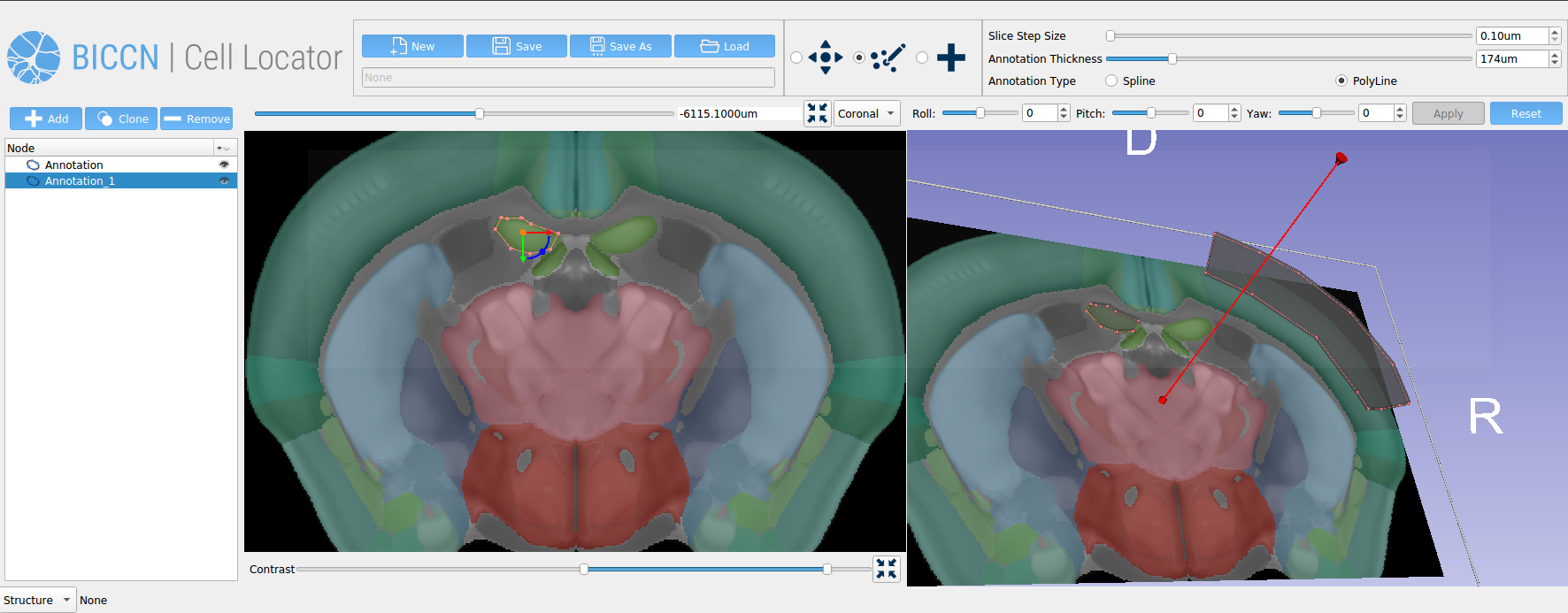
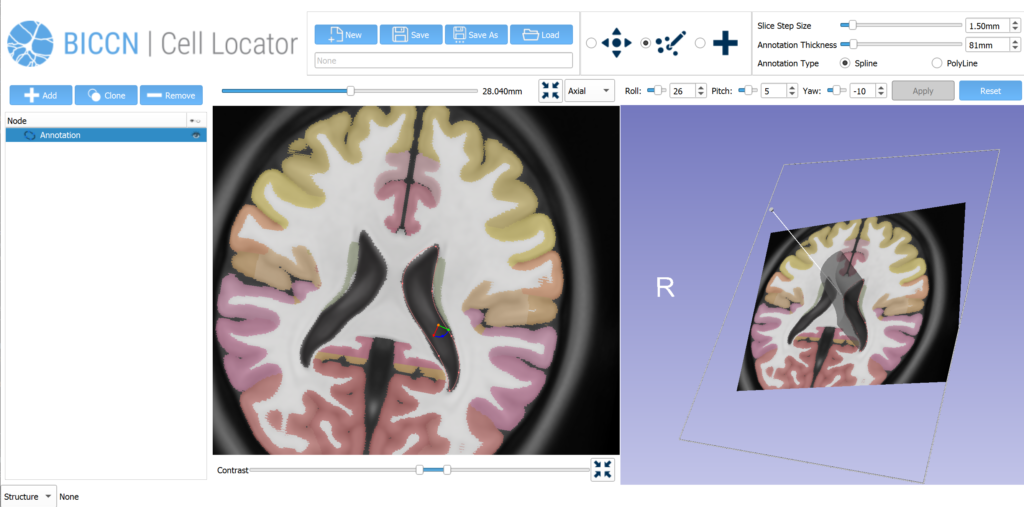
Cell Locator is an annotation tool facilitating locating brain samples in 3D reference atlases that we have developed for our partners at the Allen Institute for Brain Science and BRAIN Initiative Cell Census Network (BICCN) (Figure 1). It is an open-source cross-platform desktop application based on 3D Slicer which integrates with the Allen Institute’s workflow and web services to support neuroscience research. Here, we present our software development and collaboration strategies.
Project History
3D Slicer is a desktop application available on multiple operating systems for analysis and visualization of 2D, 3D, and 4D medical images. Slicer is a full-fledged platform enabling both rapid prototyping using python scripting and the development of custom applications (see our previous blog “Creating custom applications based on 3D Slicer”). These features make 3D Slicer an excellent choice for an annotation tool like Cell Locator.Kitware and Allen Institute for Brain Science have been collaborators since 2017, when we worked together to build software quality practices and community around the NWB 2.0 ecosystem, along with visualization and analytics tools. Allen Institute for Brain Science hired Kitware to develop Cell Locator to support the NIH’s BRAIN Initiative Cell Census Network (BICCN) project, with goals of providing references of the cell types in human, mouse, and non-human primate brains. We are now entering our third phase of work together.
Example Workflow
We’ll follow the Allen Institute’s Patch-seq data generation pipeline to demonstrate Cell Locator’s use. In this pipeline, each cell is patched to characterize its intrinsic electrical properties while a dye is injected and diffuses throughout the soma, dendrites, and axon of the cell. At the end of electrophysiology recording, the cell nucleus is extracted and prepped for RNA-sequencing to obtain the cell’s gene expression profile. Meanwhile, the brain slice containing the cell is fixed, stained and mounted, and imaged at 20x resolution, followed by high-resolution imaging of the cell for morphological reconstruction and analysis. Data and instructions associated with workflow tasks (Figure 2) are orchestrated by a workflow manager interfacing the Allen Institute web-based LIMS (Laboratory Information Management System).
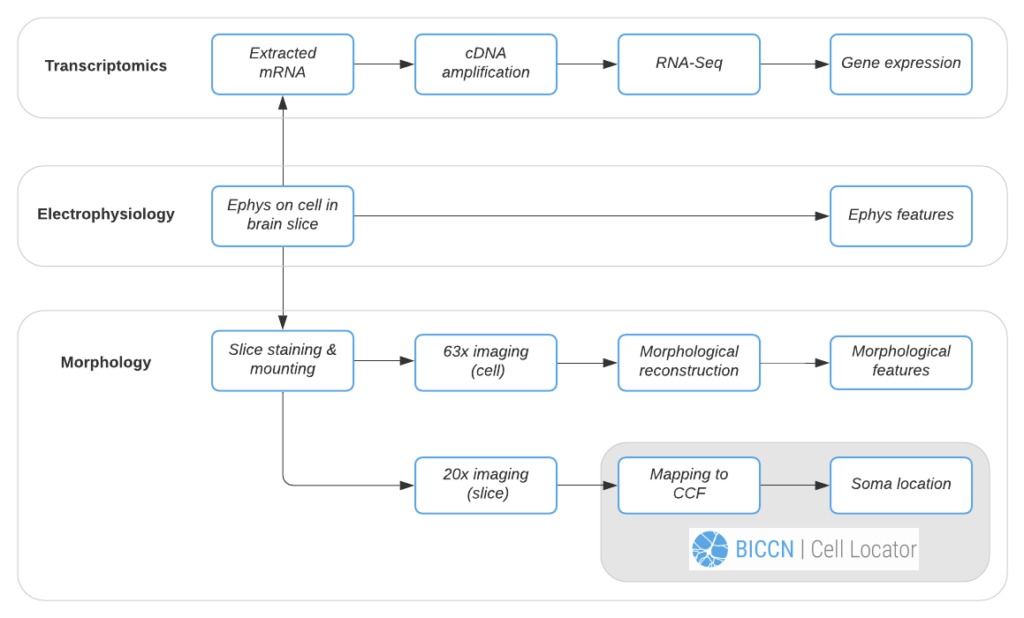
Using the 20x image of the brain slice, the operator creates a matching virtual slice in Cell Locator and annotates the cell’s soma location. The orientation of the slice and annotation coordinates are then saved and stored in LIMS. Cell Locator provides 3D navigation tools and displays the Allen Mouse Common Coordinate Framework or Allen Human Reference Atlas to assist in the alignment process. These atlases are segmented by brain structure and color-coded to help the operator correctly annotate the specimens. If the approximate location of the specimen is known in advance, as is the case with LIMS, the orientation may be supplied via command-line arguments, so that the operator would only need to make fine adjustments.
Software Quality and Best Practices
Our software development strategy in this project focused on rapid iteration and transparent collaboration with our partners at the Allen Institute, to gather feedback from users, and converge to a relevant user interface and feature set. Using GitHub issues and projects allows us to easily track the status of issues and feature requests in a Kanban board which we reviewed during our weekly team meetings. We were able to discuss these topics asynchronously in GitHub issues, and discuss implementation details in GitHub pull requests. Testing was also tracked through GitHub, with templates available for bug reports, feature requests, and QA Checklist Issues, to guide team members (Figure 3). In preparation of an upcoming release, we would evaluate the application against a release-specific checklist, and track any issues in GitHub.
Desktop/Web Integration
In the first phase of Cell Locator, the annotation file would have been saved to disk, and manually uploaded to LIMS. Now, Cell Locator can automatically push changes to LIMS through HTTP requests.As Kitware does not have direct access to LIMS (the service runs within the Allen Institute for Brain Science firewall), we instead collaborated with the Allen Institute to document the API and created a mock implementation of the API using the open-source web framework Flask. This mock server allowed us to rapidly test and verify our LIMS integration, while the LIMS API implementation itself was evolving. This strategy allowed the Allen Institute to periodically validate new features against the real LIMS service, without altering our internal development cycle.
Extensions and Future Work
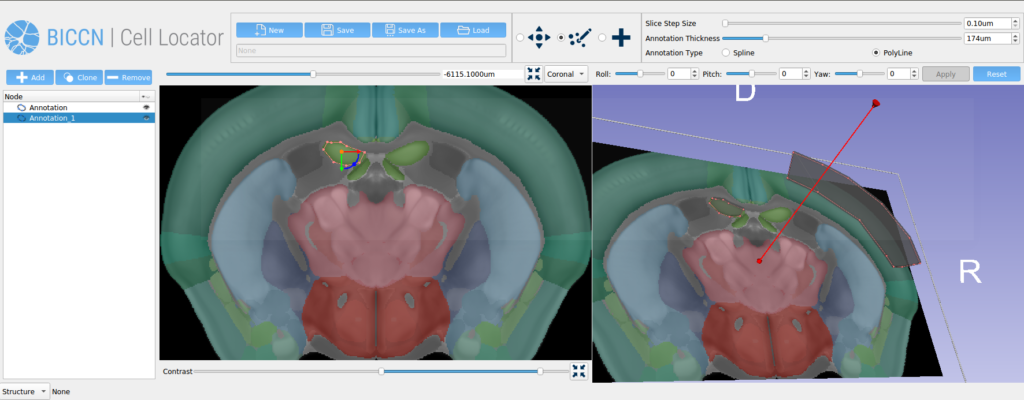
Our future work on this project will be to further develop the human pinning tool use cases, extending our phase two work that added the Allen Human Reference Atlas into Cell Locator (Figure 4). As part of the third phase of our work with Allen Institute for Brain Science, we will be engaging with the broader BICCN through GitHub and workshops, to support community needs for Cell Locator and adapt it to a wider set of lab-specific workflows.
Acknowledgments
Research reported in this publication was supported by the National Institute Of Mental Health of the National Institutes of Health under Award Numbers U01MH114812 (PI. E. Lein), U19MH114830 (PI. H. Zeng), and U24MH114827 (PIs M. Hawrylycz, L. Ng). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Research Resource Identifiers (#RRID) referenced in this publication are BICCN Cell Locator (BICCN Cell Locator, RRID:SCR_019264), BRAIN Initiative Cell Census Network (BICCN, RRID:SCR_015820), Allen Mouse Common Coordinate Framework (CCF) (Allen Mouse Brain Common Coordinate Framework, RRID:SCR_020999) and Allen Human Reference Atlas (Allen Human Reference Atlas, 3D, 2020, RRID:SCR_017764).
We would like to thank the following past and present members of the Allen Institute for Brain Science team for their contributions to this project from administration and planning, to providing early feedback and improvement suggestions: David Feng, Stephanie Mok, Nick Dee, Tamara Caper, Rachel Dalley, Arielle Leon, Rusty Mann, Zach Madigan, Rob Young, Josh Royall, Scott Daniels, Katherine Baker, Hongkui Zeng, Song-Lin Ding and Ed Lein.