Community Spotlight: Creating a Virtual Brain Atlas for Medical Education using ITK

Question: How can a pea-sized brain tumor cause such seemingly incongruous symptoms as double vision, abnormal bone growth, and infertility? Answer: Because anatomic neighbors are frequently affected by the same tumor, stroke, or injury. This is why it is crucial that medical students learn the complex spatial relationships of functionally important brain structures and pathways, most of which are not visible on the surface. After many years teaching medical neuroanatomy, we know that students find it very difficult to create a mental picture of these spatial relationships using traditional resources, such as atlases of stained-sections of brain tissue and plastic models. A few years ago, given exciting advances in 3D medical imaging, we thought surely a digital atlas of the human brain had been developed that provided direct visualization of internal brain anatomy. After an extensive search, we were surprised to find that none of the available resources met our students’ needs; in particular, all of them failed to show dozens of important structures that are too small or lack sufficient contrast to be resolved on MRI or CT scans.
It Took a Team
In retrospect, it shouldn’t have been very surprising that details were missing from digital 3D brain models, since manual segmentation of such ‘invisible’ structures requires a lot of time and patience combined with a detailed knowledge of neuroanatomy, two things that don’t often coexist in one person. We believed that a team approach could be successful, so we decided to work together to create a virtual brain atlas tailored to our curriculum and designed to help students learn human brain anatomy. Our team included a first year medical student willing to spend his summer doing manual segmentation (Joseph Rozell), a neurosurgeon (Charles Kite), a neuroanatomist (Norman Strominger), and a neuroscience educator and grant writer (Tara Lindsley). Since high resolution MRI images are essential for accurate segmentation and rendering, we obtained a head MRI of a normal volunteer on a powerful GE GENESIS-SIGNA 3 Tesla MRI, courtesy of the Neurosciences Institute at Albany Medical College.
ITK-SNAP was the Ideal Software
The team agreed that our overarching goal was to create the missing resource, study how it is used, verify its effectiveness in our curriculum, and then make it as widely available as possible. The intention to share our work led naturally to a search for open-source software. We found that ITK-SNAP had all the visualization, image processing, segmentation, and registration algorithms we needed and was easy enough to download and use, even for a team without IT support or programmers. ITK-SNAP is mainly developed by a team at the University of Pennsylvania, and is based on the ITK, VTK, and FLTK toolkits. Several other programs had similar capabilities, such as zooming and rotating the 3D models. However, several features made ITK-SNAP ideally suited for use by medical students and the way we imagined they would use the models: 1) the student could build custom models and save and share them; 2) they could ‘see through’ superficial structures to appreciate how they are spatially related to deeper structures using the opacity settings; and 3) the student could see how structures in the 3D window correspond to their appearance in MRIs in standard clinical views, making it more relevant for future physicians.
Out of 24 Million pixels, 79 Structures and Some Problems
The raw MRI data files contained 326 axial, 248 sagittal, and 300 coronal slices, which corresponded to a staggering combined total of roughly 24 million pixels. Seventy-nine structures and pathways were manually segmented in the sagittal slices by moving serially through the stack and completing one structure at a time. This was arduous and required lots of educated approximation of the location of structures not discernible on MRI. Members of the team and print atlases were consulted frequently to ensure accuracy [1,2]. A prototype of the software that used ITK-SNAP to view the image database was distributed to 135 first-year medical students and 6 faculty the following year. When we asked them about their experiences with the software, we learned that most users were thrilled with the visualization capabilities and thought the models were extremely effective for learning the spatial relationships of brain structures.
However, some students complained about the many steps that are required to download and install ITK-SNAP, and problems loading the image files. More worrisome was the feedback indicating that many users unintentionally applied edit functions with unhelpful results. Suggestions for improving the software were addressed the following summer, taking advantage of the open source nature of the software package, by creating a simplified version of ITK-SNAP [4], without the 3D rendering capability. This required adding a team member with some computing skills (Aaron Asch, an undergraduate).
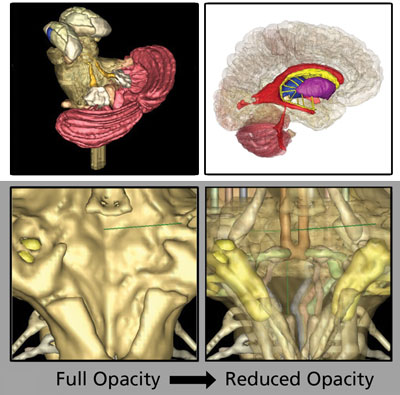
An Even Better Brain Model (v2.0)
Aaron used a CVS client and open-source utilities, such as CMake and GCC along with the Visual C++ Express IDE to compile and work with ITK-SNAP, and relied on useful instructions in the source code. The GUI editor “Fluid” from the FLTK toolkit was used to rearrange components of the user interface, adding menu buttons to simplify commonly used tasks and to introduce new educational functions. In keeping with open-source practices, this modified code was posted back to the developer’s website. Key features of this user-only version of the Virtual Brain Model (v2.0) for education are illustrated in Figure 2:
• Software binaries, image database, and instructions constitute a file size of 150 megabytes.
• Download and install takes just 2 clicks, then the software can be launched from the desktop icons.
• A simplified Main Screen retains the clinical MRI views and 3D window (Fig. 2A), but has new menu buttons, including:
• Center Axes
• Show All or Hide All structures
• Open pre-built models corresponding to specific learning objectives (Fig. 2B)
• Save Model and My Models to simplify saving custom models and accessing them later
• A Selected Structure Description window names the structure at the crosshairs (Fig. 2C).
• The structure browser includes a Groups list to select all subsets of certain larger structures (Fig. 2D).
• Easy access pull-down menus, including a step-by-step tutorial in the Help menu
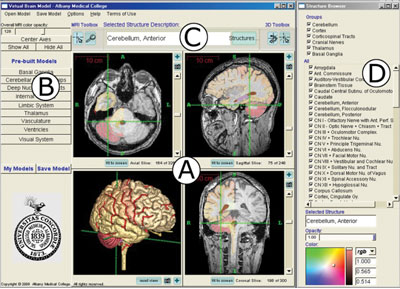
Figure 2: AMC Virtual Brain Model Software Main Screen
This version of the software has been used by three classes of medical students, and just as the original version of ITK-SNAP, it is open-source and available for download at www.amc.edu/academic/software. An abstract describing this work has been published [3], and an analysis of learning outcomes is underway.
What’s Next? Kitware on the Team
We plan to publish a paper describing the development and use this software and will submit the resource to the MedEd Portal to target its dissemination to medical and allied health professions schools. But first, we recognize a few additional changes are needed. Users around the world are asking for compatibility with other OS, an improved tutorial with guided exercises, and other changes. We are excited about the next version of the Virtual Brain Model software which will benefit from the open-source and programming expertise of Kitware. We hope the ITK open-source community will take the AMC Virtual Brain Model v2.0 for a test drive and offer us some suggestions for an even better brain.
References
[1] Miller, RA and Burack, E. Atlas of the Central Nervous System in Man. 3rd Ed. Williams & Wilkins, Baltimore, MD. 1982.
[2] Haines, DE. Neuroanatomy: An Atlas of Structures, Sections, and Systems. 6th Ed. Lippincott, Williams & Wilkins, 2004.
[3] Rozell J, Strominger NL, Lindsley TA, Kite CH. Program # 23.5 2009 Neuroscience Meeting Planner. Chicago, IL:
Society for Neuroscience, 2009. Online.
[4] ITK-SNAP: http://itksnap.org/pmwiki/pmwiki.php?n=Main.Credits
 Tara Lindsley is a Professor and the Thelma P. Lally Chair of Neuroscience Education at Albany Medical College in Albany, NY. She is a member of a team of faculty and students developing open-source virtual human anatomy atlases for education by creating manually segmented image databases and simplified, user-only software on the ITK infrastructure. Release of virtual female pelvis model software is planned for 2013.
Tara Lindsley is a Professor and the Thelma P. Lally Chair of Neuroscience Education at Albany Medical College in Albany, NY. She is a member of a team of faculty and students developing open-source virtual human anatomy atlases for education by creating manually segmented image databases and simplified, user-only software on the ITK infrastructure. Release of virtual female pelvis model software is planned for 2013.