Fast, N-Dimensional Texture Feature Maps
Introduction
The post presents texture quantification algorithms that provide a statistical description of the local texture of a 2D, 3D or N-D image. The N-Dimensional texture maps are generated with a high performance algorithm and multi-threaded implementation in the Insight Toolkit (ITK).
The software is packaged for Python and in a 3D Slicer extension.
We used ITKTextureFeatures to characterize subchondral bone texture in temporomandibular joint (TMJ) osteoarthritis (OA). Bone is a great test example for these filters due to the rich textures present in the trabecular bone (low density bone).
Installation
Python
Python wheels allow us to easily install ITKTextureFeatures and all its dependencies in order to have this texture filters ready to use in Python.They have been generated for the three main operating systems (macOS, Linux and Windows) and three versions of Python (2.7, 3.5 and 3.6).
To install Python wheels, run the following from the command-line:
python -m pip install --upgrade pip python -m pip install itk-texturefeatures
A few Python example scripts are available to quickly get started.
3D Slicer GUI Application
The 3D Slicer BoneTextureExtension is available through the Slicer Extension index.
For detailed instruction and usage instructions, please follow this tutorial.
C++
In the upcoming ITK 4.13.0 release, the module is made available by enabling the Module_TextureFeatures CMake variable when configuring ITK’s build.
Fast
These filters are performant because they are multi-threaded, they take advantage of ITK’s fast neighborhood operators, and they re-use intermediate computations and minimize memory use.
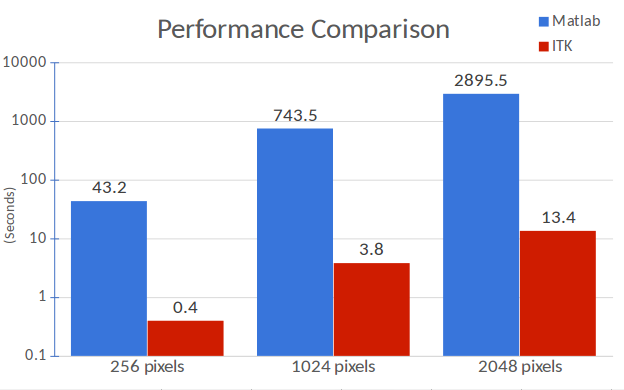
Comparisons between Matlab and ITK implementations on 2D square images of different widths (248, 1024 and 2048 pixels) demonstrate exceptional performance with the co-occurrence algorithm. Only 2D images are evaluated since Matlab only can process 2D images for this texture analysis algorithm. The ITK implementations proposed here do not share this limitation and can be used with higher dimensionality images. In addition, the Matlab implementation can only create four different texture maps (contrast, correlation, energy and homogeneity), whereas ITK can create eight (inverse difference moment, cluster shade, cluster prominence, Haralick correlation). Moreover, the iterative algorithms are optimized in our ITK implementation, and not optimized in Matlab.
N-Dimensional
These filters are the only implementation we are aware of that can compute 3D or N-D maps for these features. These maps are comprised of a collection of features which are computed per voxel. In our specific implementation, there are 18 features: 8 co-occurrence features (or Haralick features), and 10 run length features. These features are: energy, entropy, correlation, inverse difference moment (IDM), contrast, cluster shade, cluster prominence, Haralick’s correlation, short run emphasis (SRE), long run emphasis (LRE), grey level non-uniformity (GLN), run length non-uniformity (RLN), low grey level run emphasis (LGRE), high grey level run emphasis (HGRE), short run low grey level emphasis (SRLGE), short run high grey level emphasis (SRHGE), long run low grey level emphasis (LRLGE), and long run high grey level emphasis (LRHGE).

Example of texture map for LRLGE. Please note how areas the trabecular loss have high values (purple) for this feature.
Our 3D Slicer extension includes functionality to split the generated 3D textural maps with 18D features in different 3D volumes for each one of the 18 features.
More Information
- The Insight Journal article provides additional background on the theory and example application
- 3D Slicer extension
- For additional information, please contact Bea Paniagua <beatriz.paniagua@kitware.com>
Acknowledgements
This work was supported by the National Institute of Health (NIH) National Institute for Dental and Craniofacial Research (NIDCR) grant R21DE025306 (Textural Biomarkers of Arthritis for the Subchondral Bone in the Temporomandibular Joint) and NIDCR grant R01DE024450 (Quantification of 3D bony Changes in Temporomandibular Joint Osteoarthritis).
We would like to thank Dr. Larry Wolford from the Baylor University Medical Center for kindly providing the bone specimens from which we obtained the scans used in the paper. We would like to thank Drs. Lucia Cevidanes, Erika Benavides and Antonio Ruellas at the University of Michigan School of Dentistry as well, for generating the CBCT scans that are presented here.