Ginkgo CADx: Open Source DICOM CADx Environment

Ginkgo CADx [1] is a multi-platform, open core medical visualization and image-based diagnosis framework designed for specialists, practitioners, and physicians. Developed by MetaEmotion Healthcare [2], this framework provides an end-user friendly interface and extensible application for visualizing DICOM [3] studies, diagnoses, and creation of new DICOM studies.
The main input source for the application is DICOM series/studies, especially on non-radiology images based on specific workflows under IHE specifications.
Integration workflows provide formal and effective guidelines for the clinical pathway, which is the major gap we intend to fill. Without going into any technical details, a generic workflow could be summarized in five simple steps:
1. An appointment is scheduled or implicitly requested.
2. Ginkgo CADx generates or consumes the data.
3. Ginkgo CADx produces the output: Aided derived images, enhance, registration, automatic diagnose candidate or metadata.
4. The trial is reported.
5. The trial is stored and linked in the medical history.
Image integration is a mature deployment in radiology environments, where almost any image produced and consumed is integrated using HL7 and DICOM standards. Ginkgo CADx is willing to provide these workflows in other environments such as ophthalmology, dermatology, cardiology, pathological anatomy, etc.
We have developed an innovative workflow in ophthalmology diagnosis covering teleophthalmology workflows and specialist diagnosis. Advanced image processing algorithms are applied in image acquisition, including image stitching or the creation of synthetic images. The Ginkgo CADx visualization module has been extended to show all generated information in a simple way that focuses on ophthalmology.
Our image processing results are applied to dermatology samples to provide automatic skin segmentation and body surface area (BSA) calculations.
Features
The project covers these main features:
- DICOM visualization: Multiple modalities support for neurological, radiological, dermatological, ophthalmological, ultrasound, endoscopy, electrocardiogram applications, amongst others.
- DICOM communication with PACS servers.
- 3D Reconstruction: volume and surface reconstruction.
- A pluggable architecture.
Architecture
Ginkgo CADx’s backbone is a wrapped ITK and VTK streaming I/O, and the processing pipeline is adapted to the medical environment workflow. Thanks to the streaming features supported in these libraries, the application can handle large DICOM datasets with low memory requirements. A graphical UI layer has been developed using the wxWidgets library, and the DICOM backend is a wrapped DCMTK subset.
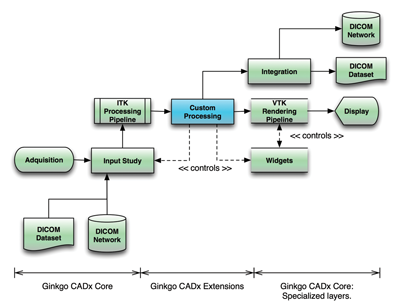 Figure 1: Framework of Ginkgo CADx
Figure 1: Framework of Ginkgo CADx
The main focus in the framework design is to wrap complexity at many levels:
- Visualization
- Interaction
- Image processing
- DICOM format and protocol
- Interoperability
- GUI
- Tasking
As these tasks are deeply specialized in heterogeneous technologies, we have been focused on providing a soft wrap over them, as well as orchestrating components interaction.
Conclusions and Future Work
Robust and standard workflows provide several improvements to diagnosis and reviewing outcomes availability; reduce the patient wait list, which results on better quality of diagnosis outcomes and economical refunds due unmissed trials; and avoid wasted time spent on tedious procedures.
The Ginkgo CADx project is constantly growing. We expect to cover more medical specialties and enhance the framework with architecture and technical improvements.
Building
Ginkgo CADx takes advantage of the CMake build system. CMake helps us to scale our build with no effort on different platforms. CMake is a great tool that lets programmers save time spent on repeating common tasks; for example, library modularization and singular settings setup. Its dependency calculator and powerful control sentences are just brilliant.
The Ginkgo CADx project is hosted at Sourceforge and uses Launchpad to support the translations platform. It is being maintained by debian-med community, bringing the binary package into debian (currently in unstable) and ubuntu (latest stable) linux-based distributions.
Acknowledgements
We wish to thank the Nuestra Señora de Sonsoles Hospital at Avila (Spain) for their funding contribution and support.
We would also like to thank Kitware’s team for developing and bringing such quality technologies and knowledge to the community.
References
[1] Ginkgo CADx. http://www.ginkgo-cadx.com
[2] MetaEmotion Healthcare: http://healthcare.metaemotion.com
[3] DICOM. http://medical.nema.org/
 Carlos Barrales Ruiz is a Senior Medical Computing Consultant at MetaEmotion specializing in medical integration standards, computational geometry, and visualization. He leads the Ginkgo CADx and HydraDICOM development. His interests are scientific visualization and geometry processing.
Carlos Barrales Ruiz is a Senior Medical Computing Consultant at MetaEmotion specializing in medical integration standards, computational geometry, and visualization. He leads the Ginkgo CADx and HydraDICOM development. His interests are scientific visualization and geometry processing.
 Javier Tovar Velasco is a Computing Engineer working in MetaEmotion’s healthcare department, taking part in the development of the open-source medical visualizer, Ginkgo CADx. He specializes in image analysis, and focuses his research on computer-aided diagnosis based on medical images.
Javier Tovar Velasco is a Computing Engineer working in MetaEmotion’s healthcare department, taking part in the development of the open-source medical visualizer, Ginkgo CADx. He specializes in image analysis, and focuses his research on computer-aided diagnosis based on medical images.
 Diego García Morate is a Computer Engineer working for MetaEmotion as CEO. He specializes in computer vision and data mining, and focuses his research on computer-aided diagnosis algorithms.
Diego García Morate is a Computer Engineer working for MetaEmotion as CEO. He specializes in computer vision and data mining, and focuses his research on computer-aided diagnosis algorithms.
When i build ginkgo cadx src but at the time of build then it give error: INCLUDE could not load file WinPackage so how to resolve this error