Integration of 3D Slicer with SOFA: An open-source pipeline for end-to-end medical simulation research

This article presents an integrated, open-source pipeline for medical simulation research. The pipeline integrates 3D Slicer [1], a state-of-the-art image analysis application, with SOFA [2], an open source framework targeted at real-time simulation. Using the cross-platform build tool, CMake [3], we integrated these two powerful frameworks to develop an application for anatomical model posing and morphing.
Introduction
3D Slicer is a free, open-source C++ software application widely used in the medical computing research community. Slicer was developed, in part, by Kitware, and it utilizes our software processes. Additionally, Slicer is built upon VTK and ITK for visualization and image processing. Slicer facilitates the clinical translation of medical computing research ideas by providing developers with an easy to use framework for image analysis and visualization. Slicer also provides a template framework that allows for the creation of customized and sophisticated user interfaces.
SOFA is a C++ library for rapid prototyping of bio-mechanical simulation applications. Integration of 3D Slicer and SOFA enables a powerful pipeline to rapidly prototype end-to-end simulation applications. This article describes how this integration was accomplished with relative ease.
Method
We employ CMake’s external projects module to download, configure, and compile the SOFA library as part of Slicer’s build process. Broadly, CMake external project allows you to pull a project’s source code without packaging it as part of the main project. The ExternalProject_Add function makes it possible to set the directives to download the project from a URL, run its configure step, and then build and install it with just the addition of a few lines of code into the CMakeLists.txt file.
SOFA’s external project command is listed below.
include(ExternalProject)
set(TAG “b79944acb7b6d7cea074db7321068f5265c2bde9”)
…
ExternalProject_Add(SOFA
SOURCE_DIR SOFA
BINARY_DIR SOFA-Build
GIT_REPOSITORY “git://public.kitware.com/Bender/SOFA.git”
GIT_TAG ${TAG}
CMAKE_ARGS
${CMAKE_OSX_EXTERNAL_PROJECT_ARGS}
-DCMAKE_BUILD_TYPE:STRING=${CMAKE_BUILD_TYPE}
${SOFA_ADITIONAL_ARGS}
…
DEPENDS
${SOFA_DEPENDENCIES}
)
Demonstration Application: Bender
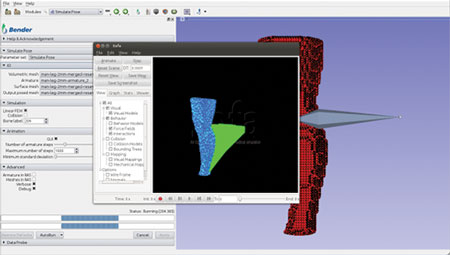
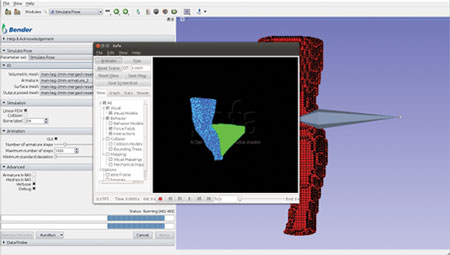
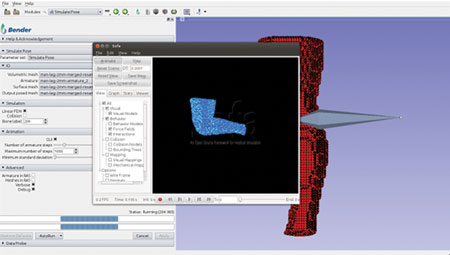
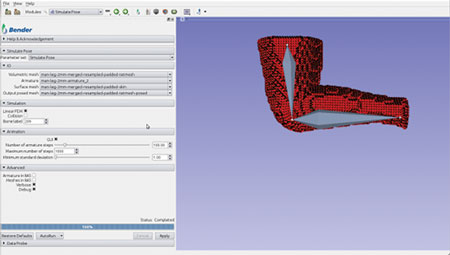
The computation pipeline consists of several Slicer modules working in unison and uses SOFA components and other libraries. This integrated pipeline was used to build an interactive software application for anatomical model repositioning, called Bender [4]. Bender allows users to change the pose of anatomical models that are represented as labeled voxel-based volumes. In order to use SOFA’s physics engine, the voxelized anatomical model must be converted into a tetrahedral mesh. To do so, we used the open-source image-to-mesh meshing library called Cleaver [5]. Cleaver was also integrated into Slicer’s build process. Figures 1-4 shows Bender’s repositioning workflow GUI and the various stages of SOFA’s simulation. The following figures illustrate Bender’s invocation of SOFA’s finite element components.
Figure 1. Initialize force loads to drive the repositioning
Figure 2. Apply forces to mesh vertices
Figure 3. Finalize and send result back to Bender
Figure 4. Update the mesh positions with the new ones computed by SOFA
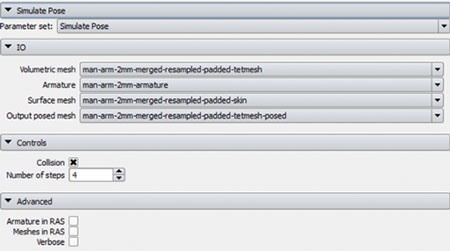
A loadable CLI module was developed to facilitate this interaction. As shown in Figure 5, the SimulatePose module provides an interface to load the required files and control the simulation within the module.
Figure 5. Simulate Pose Loadable Module
Conclusion
We presented an application that was developed by using an integrated open-source pipeline. 3D Slicer and SOFA are open-source frameworks widely used in their respective communities. The integration of 3D Slicer with SOFA provides the medical simulation community with support by two active and dynamic communities. This pipeline could potentially be used as a template platform to facilitate the development of complex medical simulations.
References
- Kitware Inc., 2014. 3DSlicer: A multi-platform free and open source package for visualization and medical image computing. Download from: http://www.slicer.org.
- Jérémie Allard, Stéphane Cotin, François Faure, Pierre-Jean Bensoussan, François Poyer, Christian Duriez, Hervé Delingette, Laurent Grisoni, et al. Sofa-an open source framework for medical simulation. In Medicine Meets Virtual Reality, MMVR 15, 2007.
- Kitware Inc., 2014. CMake: The cross-platform, open-source build system. http://cmake.org/.
- CIBC, 2014. Cleaver: A MultiMaterial Tetrahedral Meshing Library and Application. Scientific Computing and Imaging Institute (SCI), Download from: http://www.sci.utah.edu/cibc/software.html.
- Kitware Inc., 2014. Bender: A free, open source software for repositioning voxelized anatomical models. Kitware Inc. Download from: http://public.kitware.com/Wiki/Bender.
Accomplishments
The Bender development effort has been funded, in part, by the AFRL “Efficient Model Posing and Morphing Software” SBIR FA8650-13-M-6444. In addition, research reported in this publication was partially supported by the Office Of The Director, National Institutes Of Health of the National Institutes of Health under Award Number R44OD018334. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 Ricardo Ortiz is an R&D Engineer on the Medical Computing team at Kitware. He earned his Ph.D. in applied mathematics and computational sciences from the University of Iowa and served as a Postdoctoral Fellow Associate in the mathematics department at the University of North Carolina at Chapel Hill.
Ricardo Ortiz is an R&D Engineer on the Medical Computing team at Kitware. He earned his Ph.D. in applied mathematics and computational sciences from the University of Iowa and served as a Postdoctoral Fellow Associate in the mathematics department at the University of North Carolina at Chapel Hill.
 Julien Finet is an R&D Engineer at Kitware. He is involved in several medical computing projects. Julien is notably a lead developer for the Slicer, CTK, and MSVTK projects.
Julien Finet is an R&D Engineer at Kitware. He is involved in several medical computing projects. Julien is notably a lead developer for the Slicer, CTK, and MSVTK projects.
 Andinet Enquobahrie is the Assistant Director of Medical Computing at Kitware. Andinet is responsible for technical contribution and management of image-guided intervention and surgical simulation projects. His recent efforts have focused on the use of PET-CT imaging to improve the clinical effectiveness of lesion biopsy, laparoscopic surgical procedures, and tools for image-guided intervention application development and bioinformatics analysis.
Andinet Enquobahrie is the Assistant Director of Medical Computing at Kitware. Andinet is responsible for technical contribution and management of image-guided intervention and surgical simulation projects. His recent efforts have focused on the use of PET-CT imaging to improve the clinical effectiveness of lesion biopsy, laparoscopic surgical procedures, and tools for image-guided intervention application development and bioinformatics analysis.