New ITK-SNAP Features Improve Segmentation
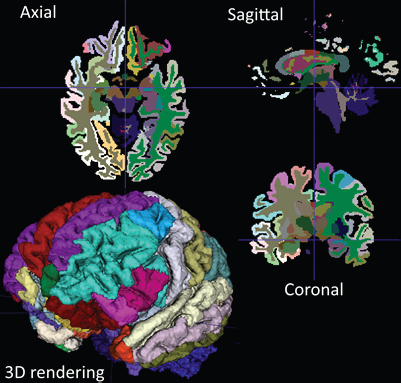
ITK-SNAP [1] is a software application that segments structures in three-dimensional (3D) biomedical images. The application is free, open source, and multiplatform. Over the last several years, the application has undergone significant refactoring and expansion. It now includes a modern, Qt based graphical user interface; extensive support for segmenting multi-modality and multi-parametric imaging datasets; and more powerful tools for performing semi-automatic segmentation. ITK-SNAP also features improved performance and scalability [2].
This article briefly describes two additions to ITK-SNAP. The first addition is run-length encoding (RLE), which compresses label maps for storing image segmentations. The second addition is a morphological contour interpolation filter, which Albu et al. first proposed [3].
RLEImage
When many adjacent pixels share the same value, as is almost always the case with label maps that represent organ segmentations, RLE can significantly save storage. Instead of keeping a line of pixels as “11111222200004555,” RLE stores the pixels as “5×1 4×2 4×0 1×4 3×5.” This reduces memory usage from 13 numbers to 10 numbers. For 3D images with millions of voxels, the savings prove far more significant.

To implement an RLE data structure for arbitrary dimensionality, ITK-SNAP relies on an existing Insight Segmentation and Registration Toolkit (ITK) class, itk::Image. ITK-SNAP implements an RLEImage of n dimensions as an itk::Image with n‑1 dimensions through the following run-length lines:
typedef std::pair<TCounter, TPixel> RLSegment; typedef std::vector<RLSegment> RLLine; typedef itk::Image<RLLine, VDimension-1> RLEImage;
For this image class to offer utility, it has to provide the access methods that ITK normally uses for the itk::Image class. These access methods include Get/Set operators as well as const iterators and writable iterators. With these operators and iterators, RLEImage can serve as a drop-in replacement for itk::Image, as long as the user code does not assume a linear pixel layout, which it frequently does.
Furthermore, to display slices, ITK-SNAP needs to extract them. Slicing serves as the most frequently used operation that involves label maps. With the release of ITK-SNAP 3.4, the switch to RLEImage only mildly affected slicing performance. In tests, slicing took about the same time with RLEImage and itk::Image. For example, it took 11.7 milliseconds to axial slice brain parcels with RLEImage, and it took 10.3 milliseconds to axial slice the same parcels with itk::Image. The worst practical case involves axial slicing a whole body that the watershed algorithm has segmented. In this case, it took 154 milliseconds to slice the body with RLEImage and 79 milliseconds to slice the body with itk::Image.
Morphological Contour Interpolation
While the use of automatic segmentation algorithms has become more popular, the medical computing community continues to measure the performance of the algorithms against a manually created “gold standard” segmentation [1]. Additionally, the community continues to use manual segmentation to train the algorithms and to solve problems for which automatic segmentation is not sufficiently reliable.
Manual segmentation, however, is often a slow and tedious process. An alternative approach to dense manual segmentation is to manually segment a sparse number of slices and use interpolation to fill in the gaps. This approach generates fully segmented volumes with much less manual input. The following screenshots demonstrate manual segmentation with the use of two labels on coronal slices.
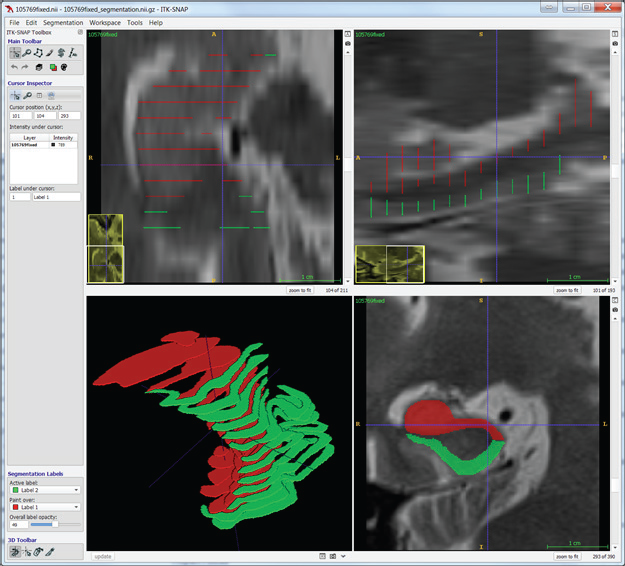
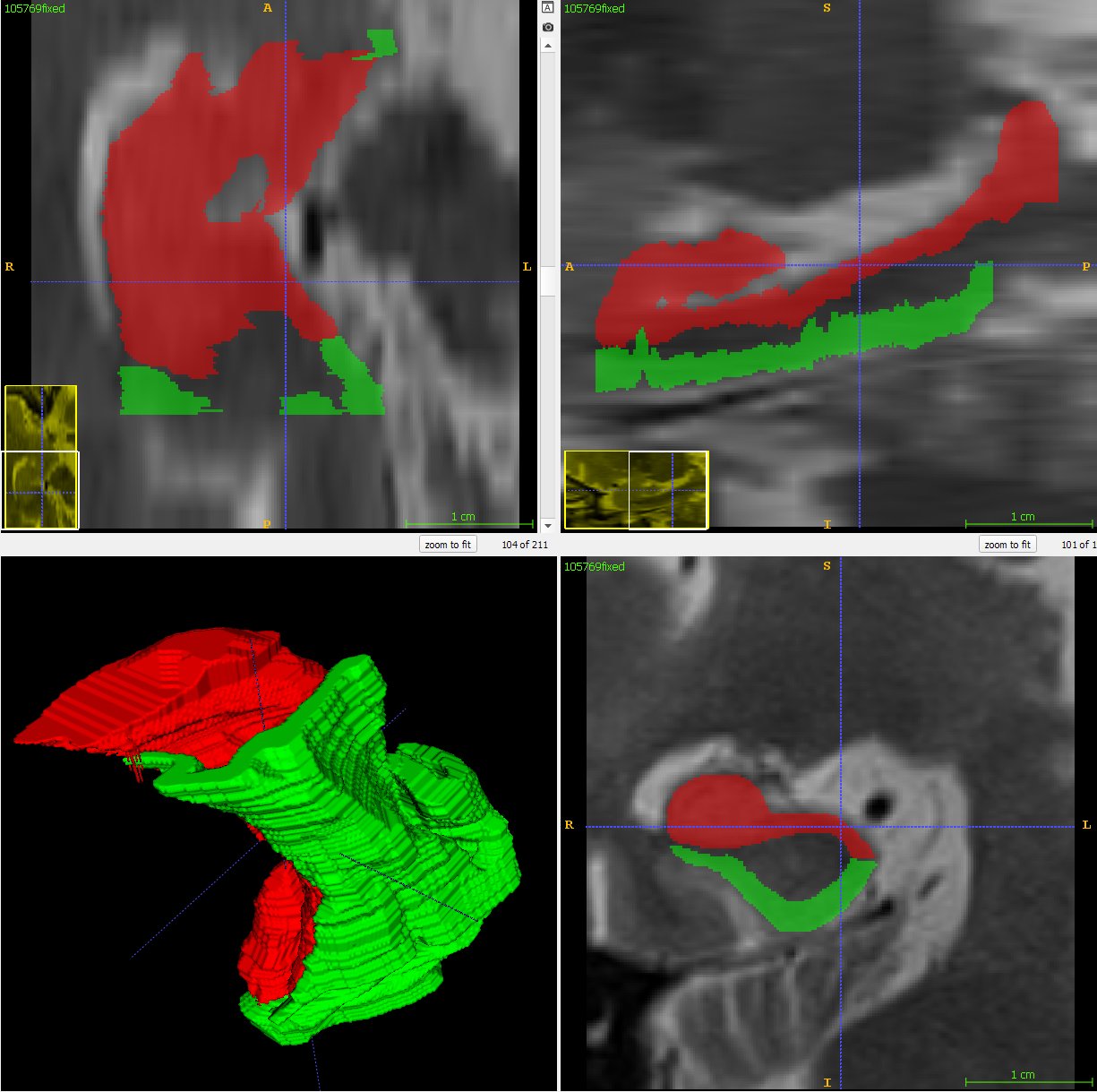
Interpolation determines the correspondence between shapes on adjacent segmented slices. To do so, it detects overlaps, aligns the overlapping shapes, generates a transition sequence of one-pixel dilations, and takes the median as the result. Recursion can aid situations in which more than one empty slice separates the original slices. Albu et al. describe the method of interpolation [3]. The method retains some distinctive features and does not suffer from excessive smoothing, unlike most shape-based methods.
The itk::MorphologicalContourInterpolator class implements interpolation. This class contains an image-to-image filter that supports 3D, four-dimensional, and higher-dimensional inputs. ITK-SNAP can restrict the filter to interpolate one label and/or to interpolate along one axis.
The below example of interpolation kept every fourth output from the original manually segmented image. The example then used the reduced segmentation as input, and it used the original segmentation as ground truth, against which it measured the interpolated segmentation.
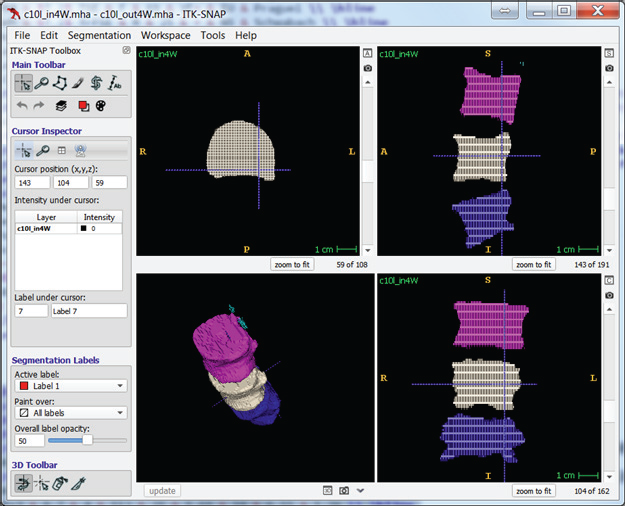
Tests on two images, “c10” and “c2,” with full manual segmentations evaluated interpolation performance. The tests left every nth slice to simulate sparse manual segmentation and cleared the rest of the images. Each test used the slices as input to the filter and then compared the output with the original image.
Dice similarity coefficient (DSC), positive predictive value (PPV), and negative predictive value (NPV) plots demonstrate the loss of precision that occurs with increasingly sparse manual segmentations. To calculate PPV and NPV, the tests assumed a background equal in size to the objects.
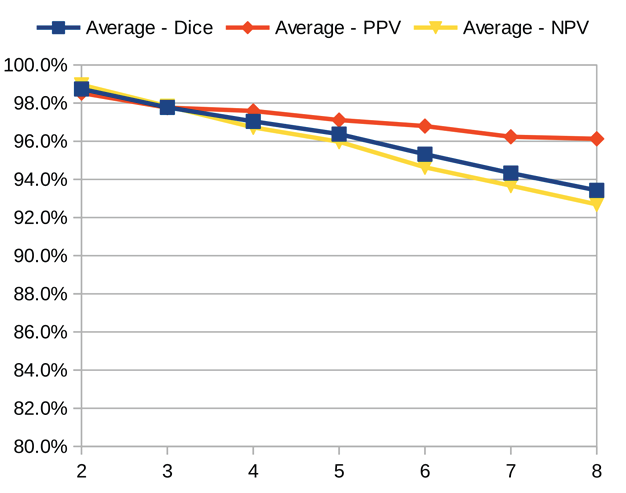
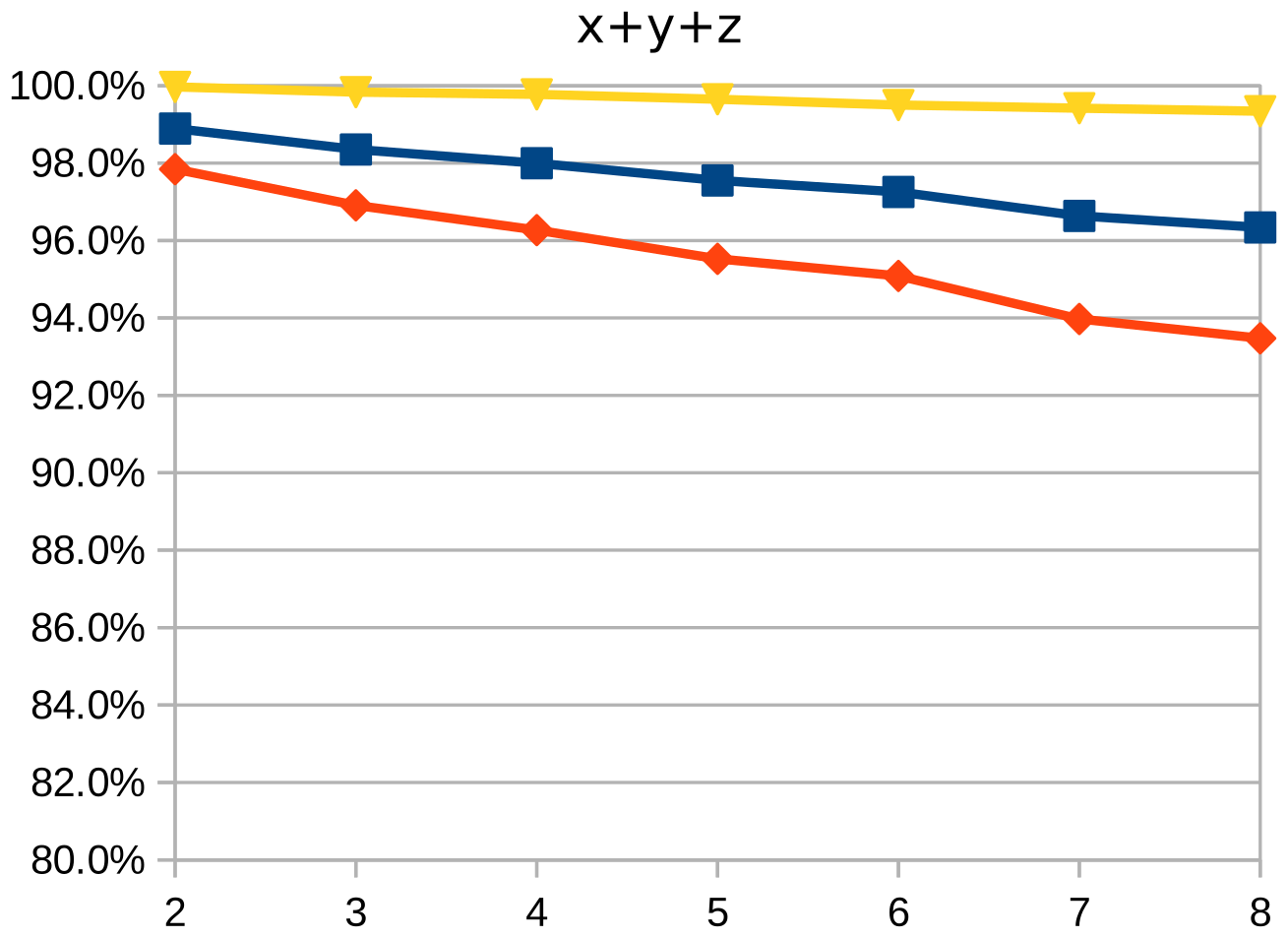
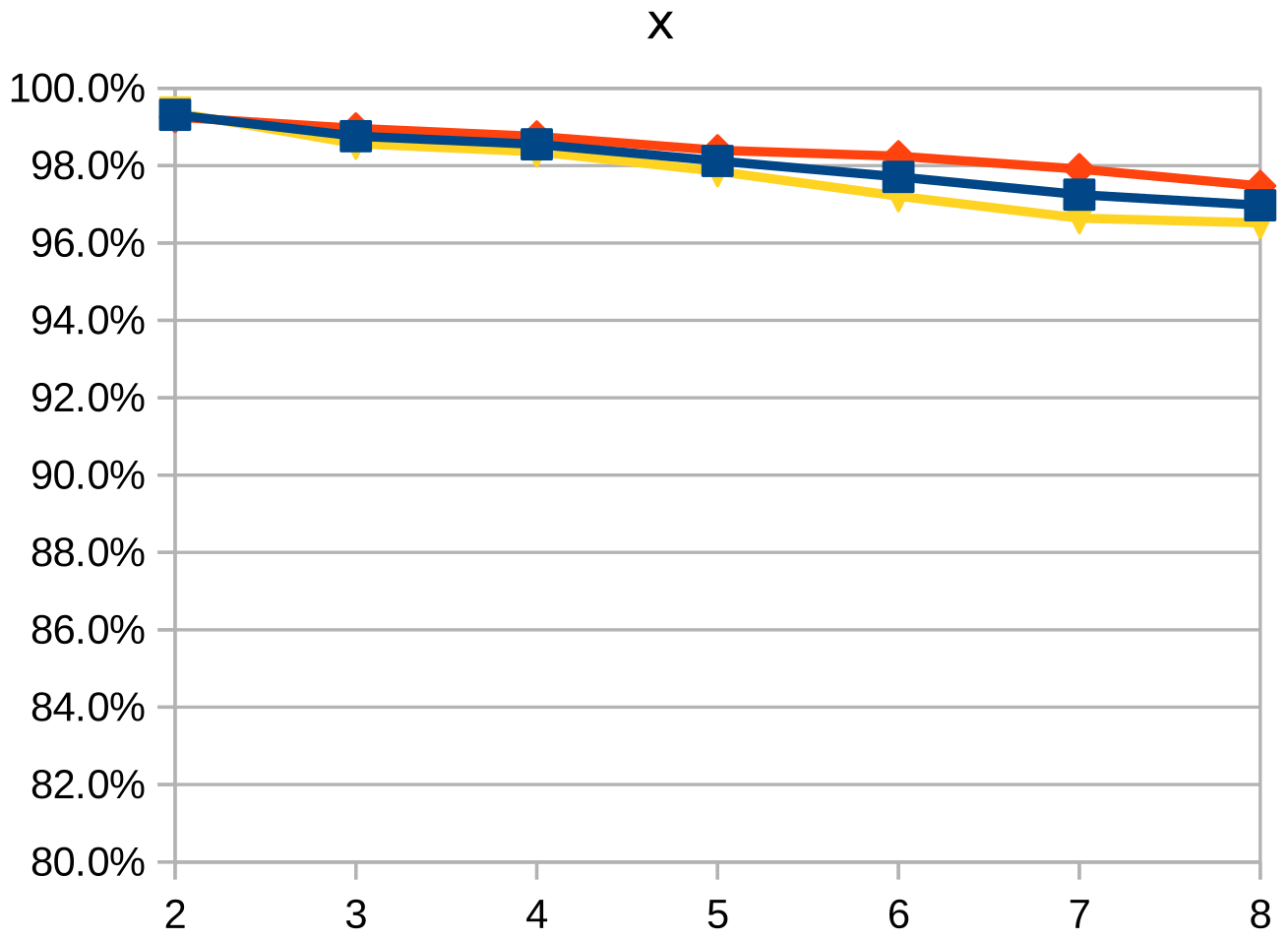
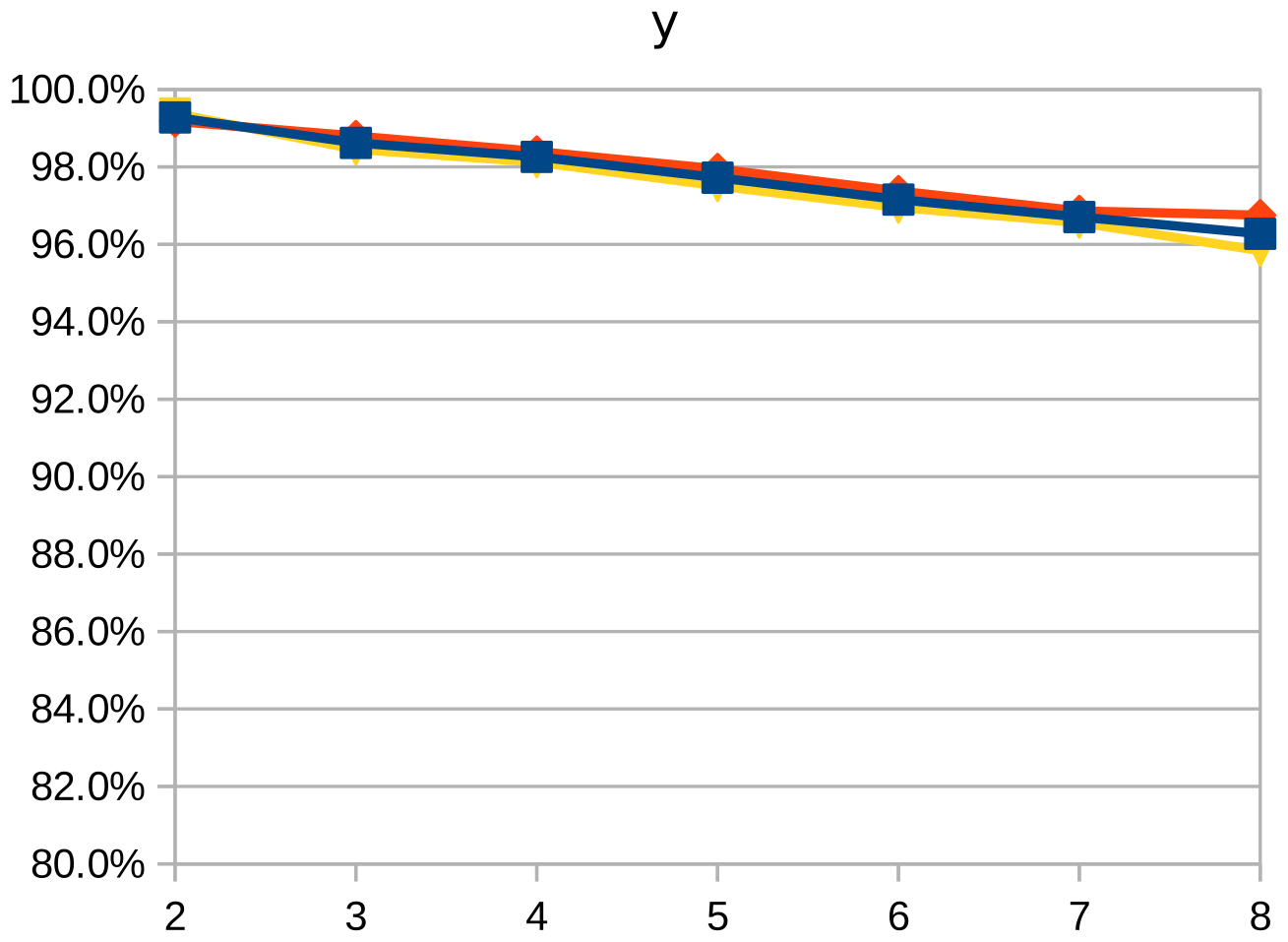
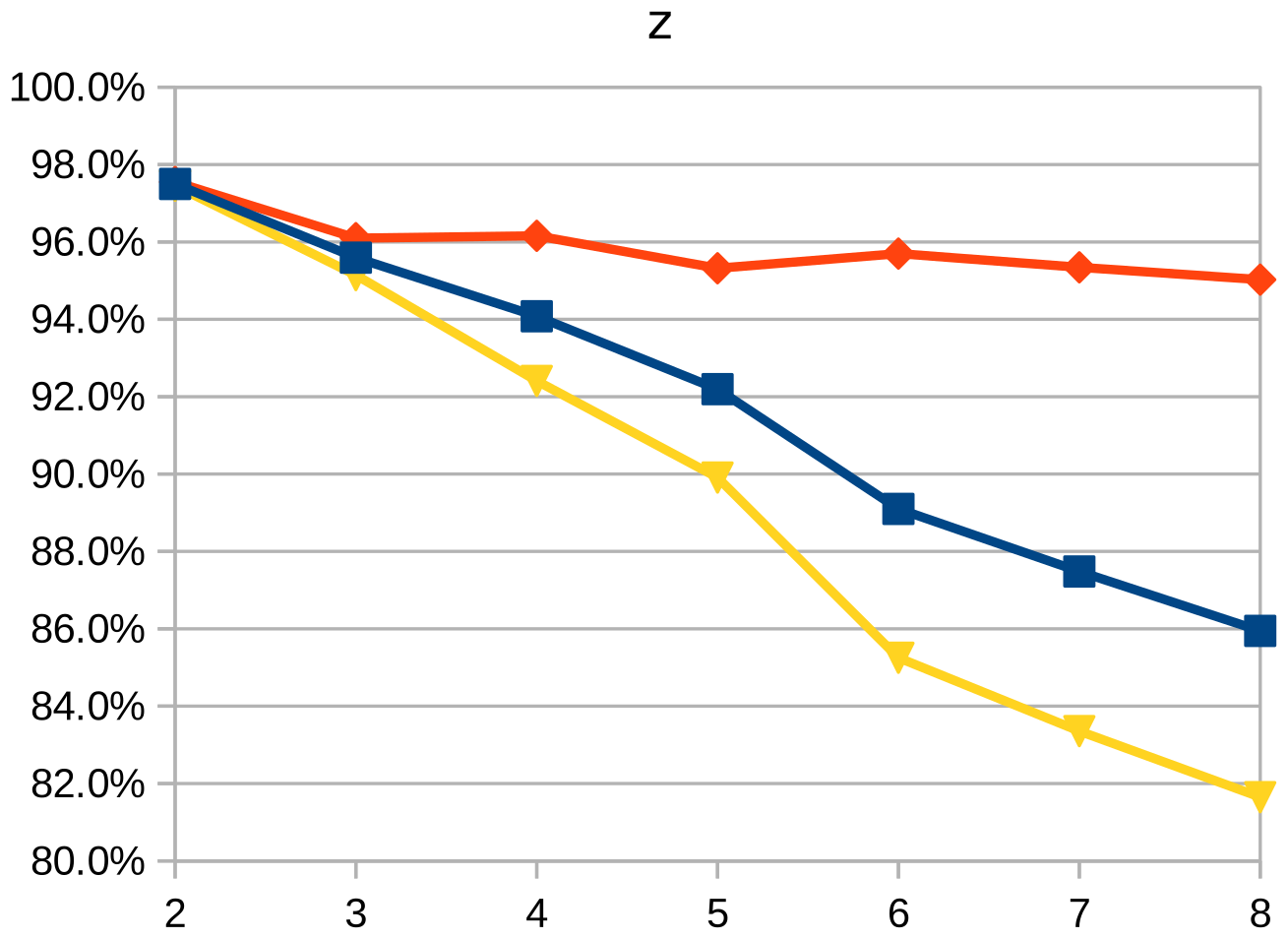
As the resolution was already three times lower along the z-axis, adding sparsity exacerbated the problem. Most errors denote false negatives, which indicate places where the interpolated image presented pixels, but the ground truth did not. Other errors denote false negatives, which indicate places where the ground truth presented pixels, but the interpolated image did not. The below screenshot represents false positives in cyan, yellow, and green, and it portrays false negatives in violet, red, and blue.
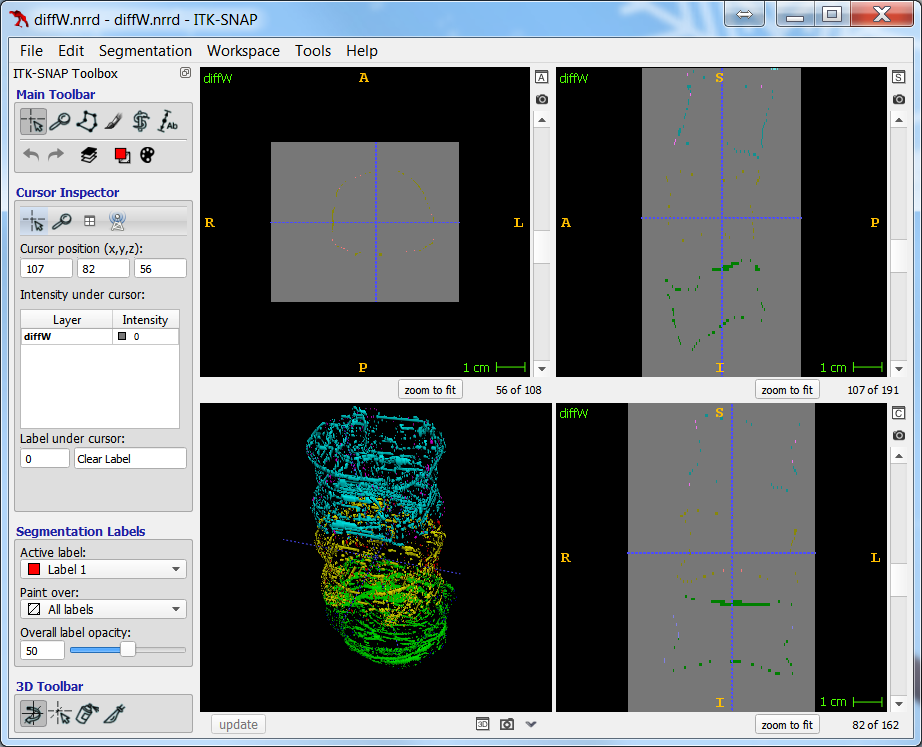
The test for the above screenshot used the same portion of “c10” that the test for the previous screenshot used.
Open Science
The general public can access the RLEIMage class and the morphological contour interpolation filter as ITK remote modules under the Apache 2.0 license. These remote modules contain source code, unit tests, test data, and documentation. To download the remote modules, please go to the following websites: https://github.com/KitwareMedical/ITKRLEImage and https://github.com/KitwareMedical/ITKMorphologicalContourInterpolation.
Acknowledgment
The National Institutes of Health grant R01-EB014346, “Continued development and maintenance of the ITK-SNAP 3D image segmentation software,” supported the ITK-SNAP segmentation work.
References
- Yushkevich, Paul A., Joseph Piven, Heather Cody Hazlett, Rachel Gimpel Smith, Sean Ho, James C. Gee, and Guido Gerig. “User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability.” Neuroimage 31(2006): 1116−1128.
2. Yushkevich, Paul A., Yang Gao, and Guido Gerig. “ITK-SNAP: an interactive tool for semi-automatic segmentation of multi-modality biomedical images.” In EMBC 2016 Proceedings, 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Orlando, Florida, 2016. New York, New York: Institute of Electrical and Electronics Engineers.
3. Albu, Alexandra Branzan, Trevor Beugeling, and Denis Laurendeau. “A morphology-based approach for interslice interpolation of anatomical slices from volumetric images.” IEEE Transactions on Biomedical Engineering 55(2008): 2022−2038.
 Dženan Zukic is a senior research and development engineer on the medical computing team at Kitware. For his dissertation, he studied the segmentation of vertebral bodies in magnetic resonance images.
Dženan Zukic is a senior research and development engineer on the medical computing team at Kitware. For his dissertation, he studied the segmentation of vertebral bodies in magnetic resonance images.
 Matthew McCormick, a technical expert at Kitware, is a major contributor to and the current maintainer of ITK. His general interests include image processing and emphasize scientific reproducibility.
Matthew McCormick, a technical expert at Kitware, is a major contributor to and the current maintainer of ITK. His general interests include image processing and emphasize scientific reproducibility.
 Jared Vicory is a research and development engineer at Kitware. While working toward his doctoral degree, he developed a method for interpolating sparse contours into a dense 3D surface.
Jared Vicory is a research and development engineer at Kitware. While working toward his doctoral degree, he developed a method for interpolating sparse contours into a dense 3D surface.
 Guido Gerig is an institute professor at the New York University Tandon School of Engineering. His work revolves around image analysis in medicine, engineering, and statistics.
Guido Gerig is an institute professor at the New York University Tandon School of Engineering. His work revolves around image analysis in medicine, engineering, and statistics.
 Paul Yushkevich is an associate professor in the department of radiology at the University of Pennsylvania. He develops novel computational methodologies for the analysis of biomedical imaging data.
Paul Yushkevich is an associate professor in the department of radiology at the University of Pennsylvania. He develops novel computational methodologies for the analysis of biomedical imaging data.