Visualizing Biological Data (VIZBI) 2015

From March 23 to March 27, 2015, I attended the VizBi conference at the Broad Institute of MIT and Harvard in Cambridge, MA. The conference featured the latest in visualizations in the life sciences community, including some impressive molecular animations.
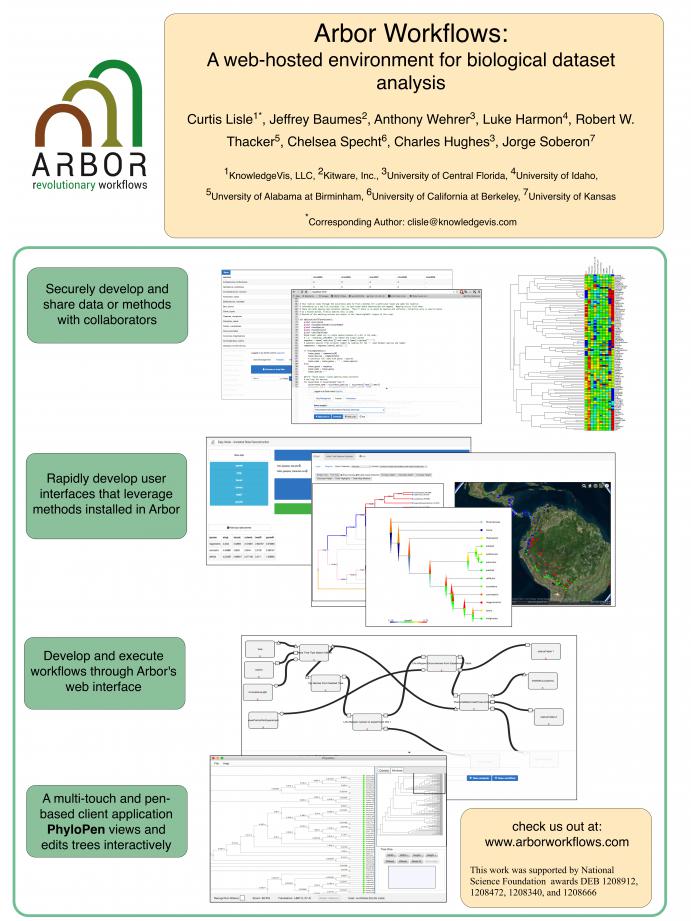
At the conference, I presented a poster (along with Curtis Lisle, Anthony Wehrer, Luke Harmon, Robert W. Thacker, Chelsea Specht, Charles Hughes, and Jorge Soberon) on the Arbor Workflows application for the Arbor phylogenetics project. Arbor uses TangeloHub to host, edit, and share data, analyses, and visualizations in the cloud. It's current capabilities revolve around phylogenetics use cases.
Some additional highlights from presentations and tutorials at the event include:
-
Galaxy is another workflow framework that has made inroads into the life sciences communities. Some initial differentiators from our poster are that TangeloHub allows cross-language workflows (Python/R) and online analysis creation and editing.
-
Cytoscape is an application for node-link views mainly focused on relating metabolites, genes, and proteins. The code is LGPL and written in Java. The tutorial made good use of Docker and iPython to demonstrate its capabilities.
-
BioJS is a loose federation of npm packages for web visualization of life science data. It is an intriguing way to incorporate a diverse set of packages, and the community has gained some traction. The most interesting points of the tutorial were the use of Cloud9 for a sandboxed coding environment, the use of browserify and parcelify for JavaScript and CSS bundling, and the slide on npm package explosion compared to other packaging systems. The Go language is also seeing rapid growth.
-
John Stasko was the keynote speaker and discussed the two main uses of visualization: exploration and presentation. I had the oportunity to speak with John at length about the future directions of his visualization team.
-
John Stone demonstrated the latest from VMD, which includes a new GPU raycaster for molecular data.
-
Jer Thorpe gave a rousing talk on visualization, focusing on the need for engaging and informative visualizations, which he termed the “OOH” and “AAH” effects. One example of this is his experience with visualizing geolocalized Twitter data.