Simulated Diffusion-Weighted Imaging

Abstract
A recent article by Van Hecke et al. [3] describes a framework for creating simulated diffusion-weighted images (DWI) for algorithmic evaluation purposes. This framework permits the modeling of intersubject variability, regional pathology, and noise. This article describes our ITK implementation of this work, which we provide to the community as open source.
Introduction
The popularity of diffusion tensor imaging (DTI) has resulted in a proliferation of corresponding analysis algorithms. The need for an evaluative framework for such algorithms motivated the developments of Van Hecke and colleagues in a recent paper [3] that encompasses modeling intersubject variability, regional pathology, and noise in creating tailored DWI from an existing DTI cohort. It should be noted that where Van Hecke’s paper also describes the registration processes to both align the DTI cohort to the common reference space and back to the individual subject space, we omit that part of the framework in this submission and allow the user to select their preferred method of image normalization. In addition, the user will need to reconstruct the resulting DTI from the simulated DWI, which can easily be performed with several publicly-available packages such as Camino [2].
Overview of Simulated DWI Creation
In addition to the modeling described above, the DWI simulation code also allows for the creation of a control group and an experimental group consisting of an arbitrary number of directional image sets, which can then be fitted to create individual DTI.
Intersubject variability is modeled using an eigendecomposition of the aligned sample DTI cohort. Specifically, for each aligned DTI, the first (i.e. longitudinal) eigenvalue and the average of the second and third (i.e. transverse) eigenvalues of each voxel within a specified brain mask are concatenated within a single vector. The matrix constituting the grouping of image vectors of eigenvalues is then decomposed to create the projections describing the variation found within the aligned cohort. These projections are used to add random intersubject variations to each of the control and experimental subject images.
Based on recent literature, pathology is introduced into the cohort using a simultaneous decrease in the longitudinal eigenvalues and an increase in the transverse eigenvalues. In propagating the change in transverse diffusion, we enforce that the ratio of the second to the third eigenvalues be the same before and after a transverse diffusion decrease.
Finally, the application of standard Rician noise is included as a possibility for enhancing the realism of the simulated DWI data. For further details, we encourage the reader to consult Van Hecke’s paper on which this work is based.
Implementation
This work was originally developed within our Advanced Normalization Tools software package and uses our ANTs command line classes. After compilation, one can invoke the help menu (by typing “CreateDTICohort –help” at the command line), which will list the following set of options and corresponding descriptions:
a, –dti-atlas inputDTIAtlasFileName
A diffusion tensor atlas image is necessary input for creating the cohort.
-x, –label-mask-image maskImageFileName
lowerThresholdValue
A mask image can be specified to determine the region(s) to which the simulated pathology operations are applied. See also the option ‘–pathology’. If no mask is specified, one is created by thresholding the atlas FA map at 0.2 unless a lower threshold is specified.
-n, –noise-sigma <noiseSigma=18>
This parameter characterizes the Rician noise in the original DWIimages. Van Hecke uses the noise-estimation method of Sijbers et al. “Automatic estimation of the noise variance from the histogram of a magnetic resonance image”, Phys. Med. Biol. 52:1335-1348, 2007.
-p, –pathology label[<percentageChangeEig1=0.05>,
<percentageChange
AvgEig2&3=0.05>,
<numberOfVoxels=
all or percentageOfvoxels >]
Users can specify the simulated pathology in a given area using a label mask. If no label is prepended to parameters, the specified parameters are applied to all labels. Pathology is simulated by changing the eigenvalues. This typically involves a decrease in the largest eigenvalue and an increase in the average of the remaining eigenvalues. Change is specified as a percentage of the current eigenvalues. However, care is taken to ensure that diffusion direction does not change. Additionally, one can specify the number of voxels affected in each region, or specify the percentage of voxels affected. The default is to change all voxels. Note that the percentages must be specified in the range [0,1]. For dimension=3, where the average transverse diffusion eigenvalues are altered, this change is propagated to the distinct eigenvalues by forcing the ratio to be the same before the change.
-w, –dwi-parameters [B0Image,directionFile,bvalue]
[B0Image,schemeFile]
This option specifies the parameters of the output diffusion-weighted images including the directions and b-values. The directions are specified using a direction file which has as its first line the number of directions. Each successive three lines contains the x, y, and z directions, respectively, and a single b-value. Note that several direction files of this format are distributed with the Camino DTI toolkit. Alternatively, one can specify a scheme file where each direction is specified, followed by a b-value for that direction, i.e. <x1> <y1> <z1> <bvalue1> … <xN><yN><zN><bvalueN>.
-r, –registered-population
textFileWithFileNames.txt
If one wants to introduce inter-subject variability, a registered DTI population to the DTI atlas is required. This variability is modeled by a PCA decomposition on a combination of the first eigenvalue image and the average of the second and third eigenvalues. The registered image file names are specified using a text file where each line is the name of an individual DTI.
-o, –output [outputDirectory ,
fileNameSeriesRootName ,
<numberOfControls=10>,
<numberOfExperimentals=10>]
The output consists of a set of diffusion-weighted images for each subject. Each file name is prepended with the word ‘Control’ or ‘Experimental’. The number of control and experimental subjects can be also be specified on the command line. The default is 10 for each group.
-h, –help
Print short/long help menus.
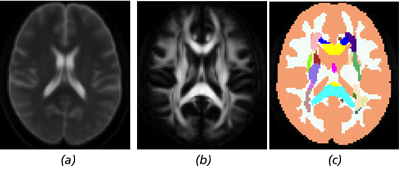
Figure 1 Axial slices of the included images: (a) the B0 image created by averaging the individual B0 images of the
NKI/Rockland INDI sample, (b) the FA image created from the DTI average, and (c) the label image that can be used to apply
anatomically specific pathology.
Simulated DWI Example
We have included a subsampled and aligned dataset3 derived from the NKI/Rockland INDI dataset consisting of 10 DTI from normal subjects; a B0 image (Fig. 1(a)); and a label mask (Fig. 1(c)) modified from the popular ICBM template consisting of 52 regions where the additional two regions are 1) unlabeled regions from the original ICBM mask where FA ≥ 0.2 and 2) the remainder of the brain. This allows for pathology to be individually prescribed for each labeled region. The FA image derived from the average DTI image is also given in Fig. 1(b). All images are aligned to the MNI152 template space [1].
Please see the full paper for a full example of a terminal output from a sample call to the routine CreateDTICohort contained in the script createDWI.sh, which is also included.
References
[1] D. L. Collins A. C. Evans and B. Milner. An MRI-based stereotactic atlas from 250 young normal subjects. In Society for Neuroscience Abstracts, volume 18, page 408, 1992.
[2] P. A. Cook, Y. Bai, S. Nedjati-Gilani, K. K. Seunarine, M. G. Hall, G. J. Parker, and D. C. Alexan- der. Camino: Open-source diffusion-MRI reconstruction and processing. In Proceedings of the 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, 2006.
[3] Wim Van Hecke, Jan Sijbers, Steve De Backer, Dirk Poot, Paul M Parizel, and Alexander Leemans. On the construction of a ground truth framework for evaluating voxel-based diffusion tensor MRI analysis methods. Neuroimage, 46(3):692–707, Jul 2009.
Acknowledgments:
Additional authors include Philip A. Cook, Brian B. Avants, and James R. Stone.

Nicholas J. Tustison recently transferred to the University of Virginia, where he continues to extol the virtues of ITK and open source to the heathen hoi polloi. Currently, he’s navigating the pernicious waters of methodological circularity in neuroscience in the hopes that any breakthroughs land him a prime spread in Oprah’s magazine (or at least a free burrito from Chipotle).